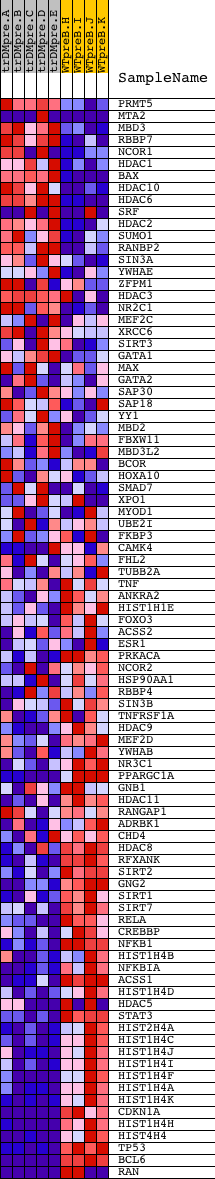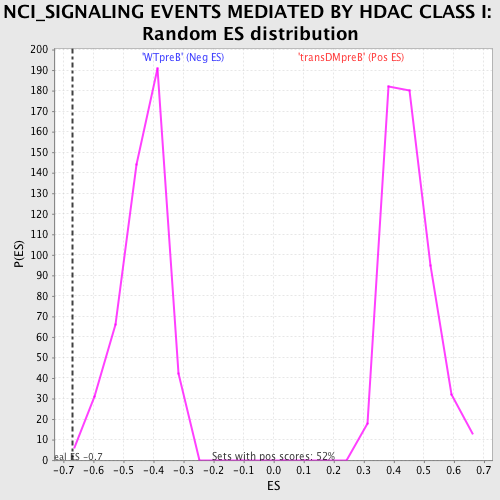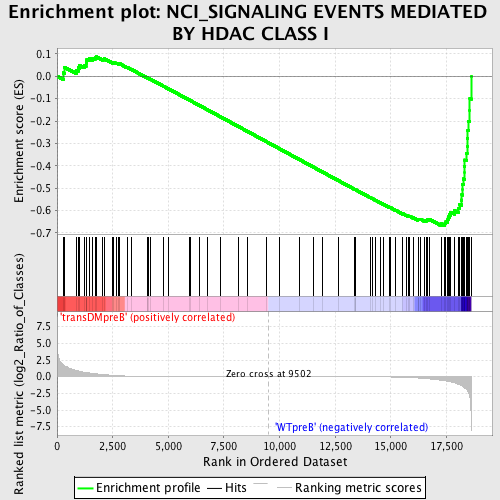
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_WTpreB.phenotype_transDMpreB_versus_WTpreB.cls #transDMpreB_versus_WTpreB |
| Phenotype | phenotype_transDMpreB_versus_WTpreB.cls#transDMpreB_versus_WTpreB |
| Upregulated in class | WTpreB |
| GeneSet | NCI_SIGNALING EVENTS MEDIATED BY HDAC CLASS I |
| Enrichment Score (ES) | -0.66864115 |
| Normalized Enrichment Score (NES) | -1.5295933 |
| Nominal p-value | 0.00625 |
| FDR q-value | 0.1352738 |
| FWER p-Value | 0.913 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PRMT5 | 6571 2817 | 277 | 1.725 | 0.0154 | No | ||
| 2 | MTA2 | 23934 18250 | 334 | 1.600 | 0.0405 | No | ||
| 3 | MBD3 | 19695 | 868 | 0.920 | 0.0279 | No | ||
| 4 | RBBP7 | 10887 6366 10886 | 950 | 0.850 | 0.0385 | No | ||
| 5 | NCOR1 | 5402 | 1004 | 0.803 | 0.0497 | No | ||
| 6 | HDAC1 | 9081 | 1222 | 0.661 | 0.0496 | No | ||
| 7 | BAX | 17832 | 1302 | 0.620 | 0.0562 | No | ||
| 8 | HDAC10 | 22165 | 1310 | 0.615 | 0.0667 | No | ||
| 9 | HDAC6 | 24197 | 1338 | 0.603 | 0.0758 | No | ||
| 10 | SRF | 22961 1597 | 1447 | 0.548 | 0.0796 | No | ||
| 11 | HDAC2 | 20055 | 1583 | 0.497 | 0.0811 | No | ||
| 12 | SUMO1 | 5826 3943 10247 | 1708 | 0.452 | 0.0823 | No | ||
| 13 | RANBP2 | 20019 | 1763 | 0.431 | 0.0870 | No | ||
| 14 | SIN3A | 5442 | 2058 | 0.325 | 0.0769 | No | ||
| 15 | YWHAE | 20776 | 2133 | 0.303 | 0.0782 | No | ||
| 16 | ZFPM1 | 18439 | 2490 | 0.199 | 0.0625 | No | ||
| 17 | HDAC3 | 4844 | 2552 | 0.185 | 0.0624 | No | ||
| 18 | NR2C1 | 10215 | 2666 | 0.162 | 0.0592 | No | ||
| 19 | MEF2C | 3204 9378 | 2762 | 0.145 | 0.0566 | No | ||
| 20 | XRCC6 | 2286 8991 | 2817 | 0.136 | 0.0561 | No | ||
| 21 | SIRT3 | 17569 3713 | 3164 | 0.086 | 0.0389 | No | ||
| 22 | GATA1 | 24196 | 3343 | 0.071 | 0.0305 | No | ||
| 23 | MAX | 21034 | 4042 | 0.036 | -0.0065 | No | ||
| 24 | GATA2 | 17370 | 4069 | 0.036 | -0.0073 | No | ||
| 25 | SAP30 | 18871 | 4117 | 0.034 | -0.0092 | No | ||
| 26 | SAP18 | 5406 | 4216 | 0.032 | -0.0139 | No | ||
| 27 | YY1 | 10371 | 4772 | 0.022 | -0.0435 | No | ||
| 28 | MBD2 | 23521 | 5025 | 0.018 | -0.0568 | No | ||
| 29 | FBXW11 | 20926 | 5928 | 0.011 | -0.1053 | No | ||
| 30 | MBD3L2 | 10638 19557 | 5980 | 0.011 | -0.1078 | No | ||
| 31 | BCOR | 12934 2561 | 6416 | 0.009 | -0.1312 | No | ||
| 32 | HOXA10 | 17145 989 | 6759 | 0.008 | -0.1495 | No | ||
| 33 | SMAD7 | 23513 | 7342 | 0.006 | -0.1808 | No | ||
| 34 | XPO1 | 4172 | 8147 | 0.003 | -0.2241 | No | ||
| 35 | MYOD1 | 18237 | 8554 | 0.002 | -0.2460 | No | ||
| 36 | UBE2I | 5819 5820 | 9396 | 0.000 | -0.2914 | No | ||
| 37 | FKBP3 | 21056 | 9999 | -0.001 | -0.3239 | No | ||
| 38 | CAMK4 | 4473 | 10887 | -0.004 | -0.3717 | No | ||
| 39 | FHL2 | 13966 | 11529 | -0.005 | -0.4062 | No | ||
| 40 | TUBB2A | 21493 | 11910 | -0.007 | -0.4266 | No | ||
| 41 | TNF | 23004 | 12661 | -0.010 | -0.4669 | No | ||
| 42 | ANKRA2 | 21578 | 13364 | -0.015 | -0.5045 | No | ||
| 43 | HIST1H1E | 21520 | 13392 | -0.015 | -0.5057 | No | ||
| 44 | FOXO3 | 19782 3402 | 13423 | -0.015 | -0.5071 | No | ||
| 45 | ACSS2 | 12242 2684 | 14100 | -0.023 | -0.5432 | No | ||
| 46 | ESR1 | 20097 4685 | 14182 | -0.025 | -0.5471 | No | ||
| 47 | PRKACA | 18549 3844 | 14320 | -0.027 | -0.5540 | No | ||
| 48 | NCOR2 | 9835 16367 | 14515 | -0.031 | -0.5640 | No | ||
| 49 | HSP90AA1 | 4883 4882 9131 | 14678 | -0.036 | -0.5721 | No | ||
| 50 | RBBP4 | 5364 | 14955 | -0.045 | -0.5862 | No | ||
| 51 | SIN3B | 5443 | 14981 | -0.046 | -0.5867 | No | ||
| 52 | TNFRSF1A | 1181 10206 | 15227 | -0.057 | -0.5989 | No | ||
| 53 | HDAC9 | 21084 2131 | 15539 | -0.081 | -0.6143 | No | ||
| 54 | MEF2D | 9379 | 15716 | -0.097 | -0.6221 | No | ||
| 55 | YWHAB | 14744 | 15786 | -0.105 | -0.6240 | No | ||
| 56 | NR3C1 | 9043 | 15860 | -0.114 | -0.6259 | No | ||
| 57 | PPARGC1A | 16533 | 16024 | -0.143 | -0.6322 | No | ||
| 58 | GNB1 | 15967 | 16221 | -0.189 | -0.6394 | No | ||
| 59 | HDAC11 | 17356 | 16254 | -0.196 | -0.6377 | No | ||
| 60 | RANGAP1 | 2180 22195 | 16328 | -0.212 | -0.6379 | No | ||
| 61 | ADRBK1 | 23960 | 16530 | -0.266 | -0.6441 | No | ||
| 62 | CHD4 | 8418 17281 4225 | 16588 | -0.283 | -0.6422 | No | ||
| 63 | HDAC8 | 24090 | 16662 | -0.303 | -0.6408 | No | ||
| 64 | RFXANK | 5378 | 16734 | -0.330 | -0.6388 | No | ||
| 65 | SIRT2 | 18318 | 17287 | -0.539 | -0.6592 | Yes | ||
| 66 | GNG2 | 4790 | 17432 | -0.617 | -0.6561 | Yes | ||
| 67 | SIRT1 | 19745 | 17477 | -0.643 | -0.6472 | Yes | ||
| 68 | SIRT7 | 9923 | 17563 | -0.694 | -0.6395 | Yes | ||
| 69 | RELA | 23783 | 17583 | -0.705 | -0.6282 | Yes | ||
| 70 | CREBBP | 22682 8783 | 17641 | -0.746 | -0.6181 | Yes | ||
| 71 | NFKB1 | 15160 | 17682 | -0.778 | -0.6066 | Yes | ||
| 72 | HIST1H4B | 6820 | 17854 | -0.907 | -0.5999 | Yes | ||
| 73 | NFKBIA | 21065 | 18048 | -1.142 | -0.5902 | Yes | ||
| 74 | ACSS1 | 12729 | 18096 | -1.212 | -0.5715 | Yes | ||
| 75 | HIST1H4D | 11410 | 18178 | -1.322 | -0.5526 | Yes | ||
| 76 | HDAC5 | 1480 20205 | 18194 | -1.345 | -0.5297 | Yes | ||
| 77 | STAT3 | 5525 9906 | 18210 | -1.378 | -0.5063 | Yes | ||
| 78 | HIST2H4A | 15240 | 18213 | -1.384 | -0.4821 | Yes | ||
| 79 | HIST1H4C | 6643 | 18272 | -1.523 | -0.4584 | Yes | ||
| 80 | HIST1H4J | 11411 | 18310 | -1.638 | -0.4316 | Yes | ||
| 81 | HIST1H4I | 6645 | 18313 | -1.645 | -0.4028 | Yes | ||
| 82 | HIST1H4F | 6644 | 18328 | -1.691 | -0.3739 | Yes | ||
| 83 | HIST1H4A | 11590 | 18396 | -1.845 | -0.3450 | Yes | ||
| 84 | HIST1H4K | 11412 | 18430 | -1.967 | -0.3122 | Yes | ||
| 85 | CDKN1A | 4511 8729 | 18439 | -2.021 | -0.2771 | Yes | ||
| 86 | HIST1H4H | 7618 | 18460 | -2.146 | -0.2405 | Yes | ||
| 87 | HIST4H4 | 6747 | 18489 | -2.351 | -0.2007 | Yes | ||
| 88 | TP53 | 20822 | 18549 | -2.941 | -0.1522 | Yes | ||
| 89 | BCL6 | 22624 | 18554 | -3.060 | -0.0986 | Yes | ||
| 90 | RAN | 5356 9691 | 18611 | -5.795 | 0.0003 | Yes |